Research
The overarching goal of our lab is to understand the interplay between human gut microbiota and host aging.
The human gut microbiota represents incredible genetic and metabolic diversity that impacts our gastrointestinal health and systemic immunity. As our microbiota alters in diversity as we age, how does it impact our health? At the community level, fecal microbiota transplantation from healthy donors improves healthspan and lifespan of aging cohorts in mouse and fish models. At the species level, probiotic supplementation extends lifespan in both C. elegans and mice.
While the gut microbiota contributes to age-associated physiology in its host organisms, we do not yet understand: 1) which gut bacterial species in the community promote healthy aging in the host, and 2) how do bacterial and host metabolic pathways play a role in this process?
We will leverage metabolomics, microbiology, and genetics to define the regulation of host physiology by the gut microbiota, at the molecular, cellular, and organismal levels. We aim to identify novel molecular mechanisms of gut microbe-host interactions and develop methods of intervention that would ameliorate age-associated health decline in the mammalian host.
Goals
1. Mechanistically characterize gut bacterial species and their bioactive molecules in host aging and physiology using genetic models
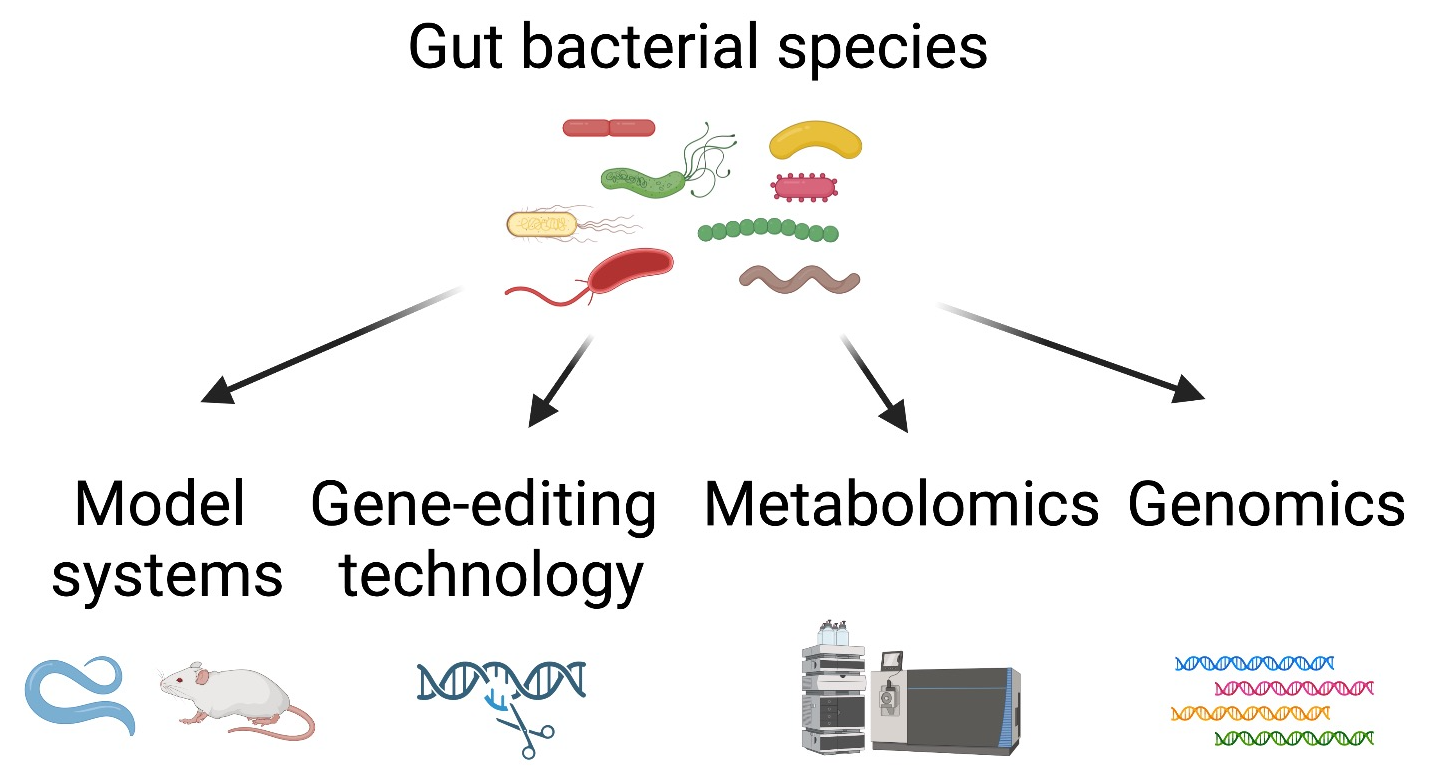
The human microbiota contains diverse bacterial phyla, encompassing remarkable genetic and metabolic diversity. However, largely due to their genetic intractability, vast majority of human gut bacteria and their bioactive molecules have not been mechanistically studied for their impact on host health and longevity.
We will address 1) which members of the gut microbiota contribute to host health and longevity, and 2) what are the bacterial and host’s genetic and metabolic pathways responsible for the host responses?
This work will identify host-beneficial gut bacteria, their metabolic pathways, and bacterially responsive pathways in the host.
2. Identify functional contributions of gut microbial communities in the aging and long-lived humans
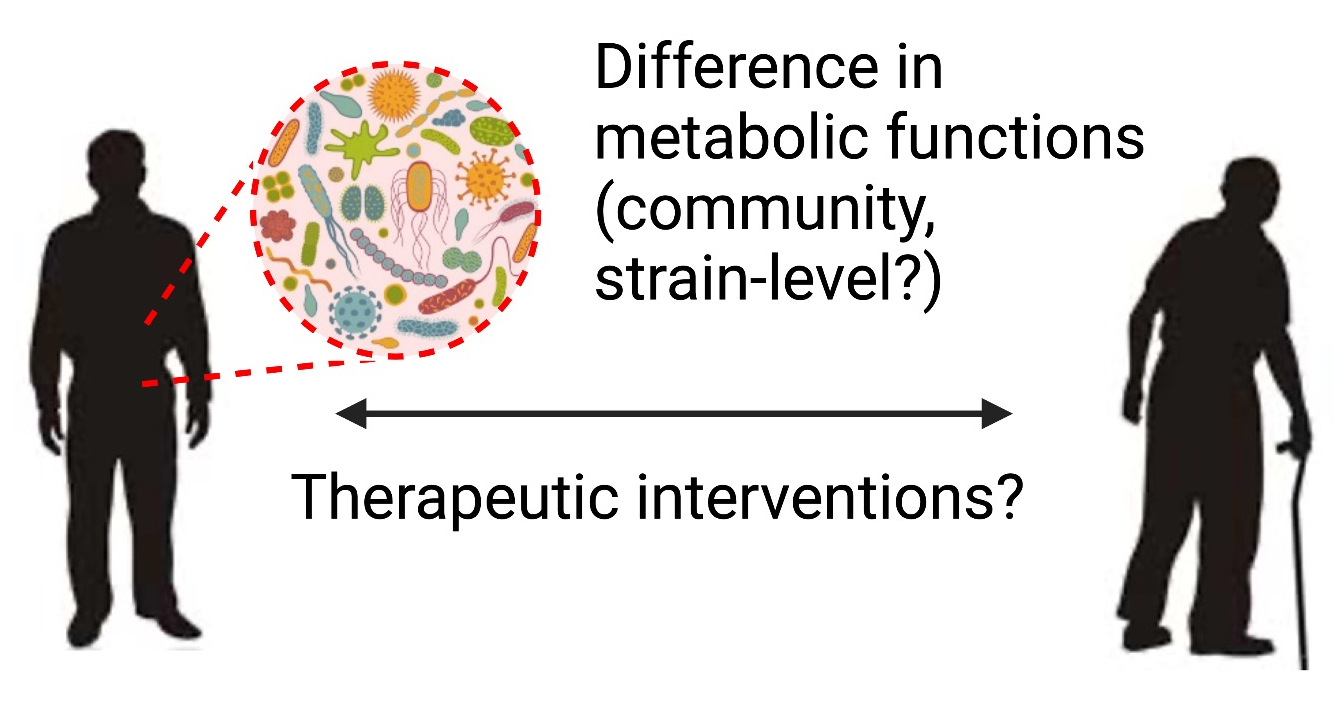
Microbiota diversity decreases with age, suggesting age-associated changes in gut microbial composition and host biological processes.
We aim to 1) identify metabolic contributions of gut microbial communities in aging and long-lived human populations, and 2) model promising metabolic phenotypes in mice for mechanistic studies.
This work will shed light on mechanistic contributions of gut bacterial species and communities to human aging and physiology.
3. Dissect the interplay between gut bacterial metabolism and host transcriptional and chromatin regulation
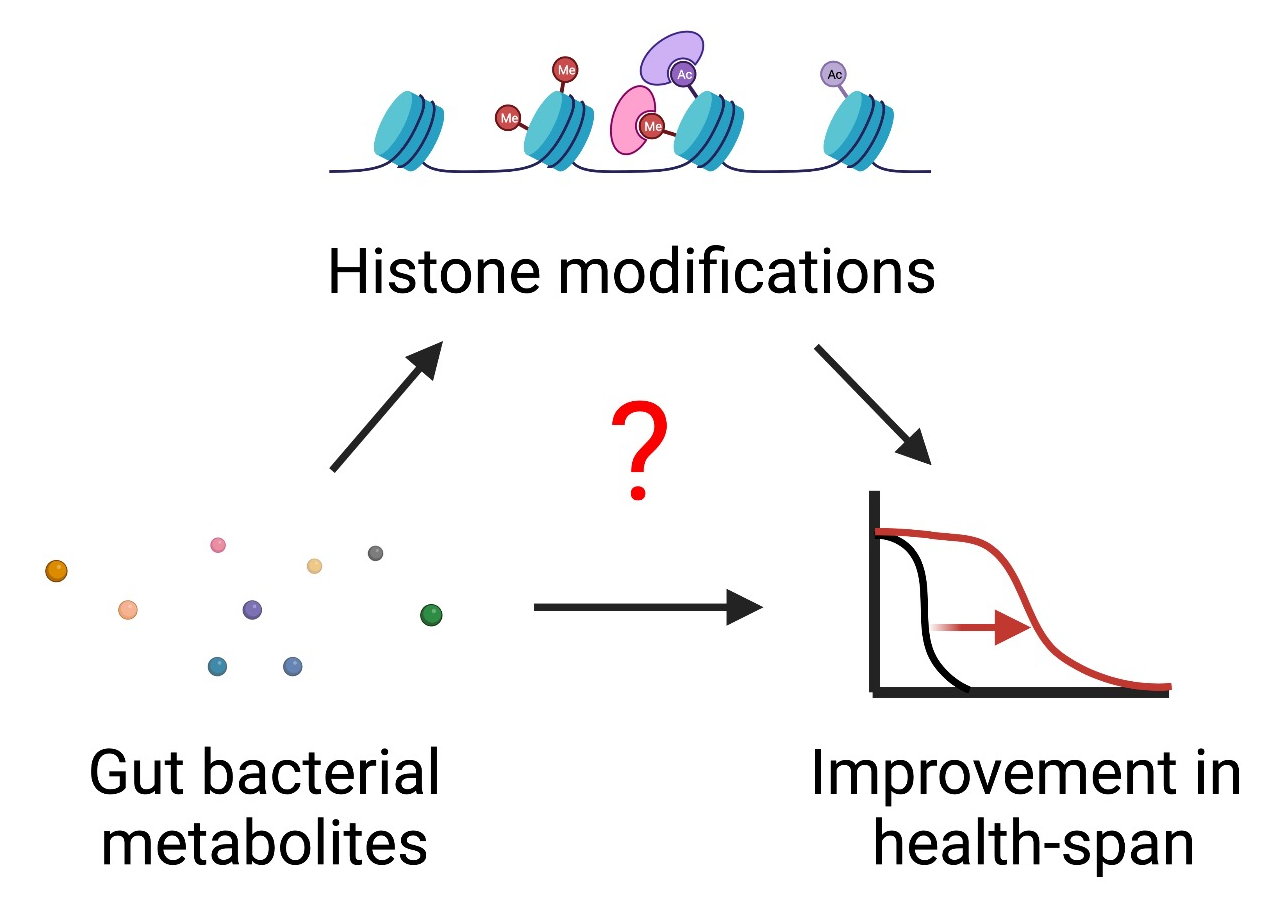
The gut microbiota’s metabolic impact on the chromatin and transcriptional regulations in distinct tissues and cells is understudied yet is emerging as a powerful mechanism of microbiota-host interactions. We recently identified a panel of the most prevalent bacterial metabolites in the human gastrointestinal tract. A few of them (e.g., butyrate, serotonin) have recently been shown to modify histones and alter transcription in distinct cell types.
We will investigate: 1) which histone modifications are responsive to gut bacterial metabolism, and 2) how do gut bacterial expression regulate host histone modifications, transcriptional programs, and cellular physiology?
This work will provide mechanistic insight into the host biology responsive to microbial signaling and uncover candidate predictors of human health and disease.